About Sanger sequencing
How does Sanger sequencing work?
Sanger sequencing, also known as the chain termination method of sequencing, is based on the selective incorporation of chain-terminating dideoxynucleotide triphosphates (ddNTPs). Various sized fragments are produced that are then separated by size during capillary electrophoresis. Sanger sequencing reactions require a double-stranded DNA (dsDNA) template (most commonly plasmids and purified PCR products), DNA polymerase, deoxynucleotide triphosphates (dNTPs), ddNTPs and primers.
The ddNTPs are particularly important due to their lack of a 3’-OH group which prevents the formation of a phosphodiester bond between two adjacent nucleotides. As such, when a ddNTP is incorporated into a DNA sequence, DNA polymerase is unable to extend the sequence past the ddNTP, terminating the reaction. In our process, the ddNTPs are fluorescently labelled to allow for the detection of terminated fragments in an automated DNA sequencing instrument.
There are 4 main steps to the Sanger Sequencing workflow:
- PCR amplification
- PCR purification
- Sequencing/capillary electrophoresis
- Data analysis
PCR Amplification: During PCR amplification, dsDNA is denatured to single-stranded DNA (ssDNA), allowing a primer to bind the template DNA next to the region of interest. DNA polymerase extends the primer by adding a single nucleotide complementary to the template, allowing for extension of the sequence one nucleotide at a time. When a ddNTP is incorporated into the sequence instead of a dNTP, the chain is terminated, which results in products of varying lengths.
PCR Purification: Amplification is followed by a purification process to removes salts, unincorporated ddNTPs, dNTPs, primers, and DNA polymerase prior to sequencing.
Sequencing: During sequencing, the extension products are injected by an electrical current into a long glass capillary filled with a gel polymer. The application of an electrical field migrates the negatively charged DNA products towards the positively charged electrode, allowing for the separation of extension products by size. When using fluorescently labelled ddNTPs on an automated instrument, as extension products migrate to the end of the capillary, a laser excites the fluorescent label at the end of the product which emits light back at a characteristic wavelength. This wavelength is detected by a light sensor, and the software translates the signal into a base call. When capillary electrophoresis complete, you will get a data file (an .ab1 file) that provides you with the DNA sequence, as well as an electropherogram.
More information about the process of Sanger sequencing can be found below, and at the following links:
- The ThermoFisher website provides an overview of the process, including an explanatory video.
- The ScienceDirect website contains a collection of articles related to Sanger sequencing.
What can Sanger sequencing be used for?
Sanger sequencing can generate a sequence of 800 or more contiguous high-resolution bases (Ewing & Green, 1998). Sanger sequencing is often used to:
- Identify single nucleotide polymorphisms (SNPs), small indels and mosaic mutations.
- Verify a cloned plasmid insert.
- Identify unknown bacterial isolates
- Confirm next-generation sequencing (NGS) variants.
- Sequence GC-rich regions poorly covered by NGS.
- Study a small subset of genes.
How can the Sequencing and Bioinformatics Consortium help me with Sanger sequencing?
We provide automated DNA sequencing using Applied Biosystems 3730XL instrumentation and chemistries. This instrument is the gold standard for high-throughput genetic analysis, allowing up to 96 samples to be sequenced in a single run.
What type of template can I submit for Sanger sequencing?
We accept the following purified templates:
- plasmid DNA
- PCR products
- cosmids
- fosmids
- BACs
More information on the types of templates you can submit, as well as the required concentrations and volumes for specific sample types can be found in our Submission Guidelines.
How much will my sequencing cost?
The total cost of your sequencing will depend on the number of samples submitted and the service option selected. Detailed information on our service options and associated pricing can be found under Services and Pricing.
When will I be invoiced? How can I pay?
We generate invoices after the close of each fiscal quarter. If you require an invoice urgently, please email us at sequencing.centre@ubc.ca. Payment from non-UBC clients can be made by cheque or wire transfer.
Can I sequence DNA that may have been stored incorrectly?
If you have any reservations about your DNA quality, we suggest running an agarose gel on your DNA to check for any signs of degradation prior to submitting to the SBC for sequencing.
How do I sequence a long amplicon or a plasmid with a large insert?
Sanger sequencing allows for the generation of a sequence of 800 or more contiguous high-resolution bases (Ewing & Green, 1998).
If there is a need to sequence a longer insert, or even your entire plasmid, proceed by one of the following options.
- If you are looking for mutations in a known sequence, you can tile the sequence of interest with individual primers.
- Use computer software to design primers that will tile the length of the DNA of interest. Ensure that your primers meet our sequencing specifications. Design your sequenced fragments to overlap, because the base calls at the beginning and end of each sequenced fragment can sometimes be of lower quality. Have all your primers synthesized at once and submit your primers along with your template for simultaneous sequencing. Remember to include enough template DNA for all required reactions.
- If you are looking to sequence an unknown region, you can ‘primer walk’ (Shapter & Waters, 2014) the sequence of interest with individual primers.
- Use computer software to design a single primer that will bind a known sequence in your DNA of interest. Ensure that your primer meets our sequencing specifications. Have your primer synthesized and submit this along with your template for sequencing.
- Based on your sequencing results, design one or more new primers that will sequence further bases along your region of interest. Have these primers synthesized and submit them along with your template for sequencing. Repeat this process until your sequencing requirements have been met.
- If you are looking to sequence an unknown insert that is greater than 10 kb in length, next-generation sequencing might be an option.
- We recommend contacting us at sanger.submissions@ubc.ca to discuss how we can help you obtain your complete sequence.
For tips on primer design, please refer to the question "Can you help me design my primers?".
Can I directly sequence a PCR product?
While it is possible to directly sequence a PCR product without first cloning the fragment, it can be tricky. The following tips will maximize your chance of sequencing success.
- Make sure you amplified the right fragment. Aside from confirming the correct size of your amplified product and the absence of amplification in a negative control, there are numerous ways to verify that you amplified the correct fragment including:
- Cutting with a restriction enzyme to verify any known restriction sites in the fragment.
- Using nested primers to reamplify the desired product to verify its identity and eliminate any spurious products.
- Remove all residual PCR primers, unincorporated nucleotides and spurious products.
- Refer to the question "How should I prepare my DNA prior to submission for sequencing?" for more information.
- Quantify your amplicon prior to submitting.
- Refer to the question "How should I quantify my DNA prior to submission?" for more information. We will adjust the amount of DNA input into the reaction depending on the amplicon length, based on the quantification information provided during submission, so it is important to quantify accurately.
- If your PCR primers will also be used as your sequencing primers, make sure they meet our primer requirements as stated in the primer recommendations in our submission guidelines.
- Note that inefficient primers may work for PCR (an exponential process), but the same primers may fail for sequencing (a linear process).
- Refer to the question "My primers produce an amplicon during PCR, but the same primers do not produce a sequence during Sanger sequencing. Why is this?" for additional information.
Can you help me design my primers?
If you are new to primer design, here are some general guidelines to follow:
- Primers are always specified 5’ to 3’, left to right. Verify that your primers are designed and ordered in the correct orientation.
- Use primer design software to avoid primers with hairpin or dimer characteristics. Thermofisher provides a free primer design tool on their website.
- Ensure that your primer is between 18 to 25 nucleotides in length, with a Tm between 55°C and 60°C.
- Ensure the 3′ end of the primer is a C or G to promote binding.
- Avoid dinucleotide repeats and runs of 3 or more of one base as this may cause primer mispriming.
- Verify the specificity of your primers to prevent binding to other non-specific regions through NCBI Primer BLAST or UCSC in-silico PCR.
- Submit custom primers in a 1.5 ml Eppendorf at a concentration of 5 pmol/µl, resuspended in distilled water or 10 mM Tris pH 7-8, with no EDTA.
What are the additives betaine and DMSO used for?
The addition of DMSO and betaine during cycle sequencing are often recommended when sequencing GC-rich samples (Choi et al., 1999). These additives help denature the DNA duplex to allow the sequencing polymerase to process the template.
What are the concerns when sequencing GC-rich samples? What should I do if my template has secondary structures?
GC-rich samples are prone to forming secondary structures such as hairpin loops. These secondary structures will prevent the sequencing polymerase from processing the template. Sequences with secondary structure may initially proceed normally but suddenly experience a drop-in signal or end prematurely.
If a secondary structure is encountered during sequencing, your primer may need to be redesigned. The position of the primer may need to be moved to the complementary strand, or to a location nearer to the secondary structure. Additions of betaine and DMSO may also assist (Choi et al., 1999). Please email us at sanger.submissions@ubc.ca if you have any concerns regarding difficult-to-sequence templates.
I’ve performed the Sanger sequencing reaction in my own lab. How do I purify the products so the electrophoresis will work?
Ethanol precipitation, dextran gel (Sephadex), and magnetic bead-based purification methods can all be successful at purifying Sanger reactions post-amplification, prior to injection on the 3730XL. The utilization of these methods will largely be determined by cost and the equipment your lab has access to. Improper purification can result in failed sequencing runs or dye blobs.
What do I need to know about submitting samples?
How should I prepare my DNA prior to submitting for sequencing?
High-quality template DNA is crucial for high-quality sequencing results. Our clients have had much success with purifying plasmid and PCR products with commercially available kits, such as those provided by QIAGEN. To optimize sequencing results, DNA should be eluted in distilled water or 10 mM Tris pH 7-8, with no EDTA. Samples that contain EDTA generally do not sequence well.
PCR products can also be run on an agarose gel to allow purification of a desired band using a gel extraction kit. Make sure that the dim background smear near the bottom of your gel, extraneous bands, primers and residual nucleotides are removed, as these will negatively affect your sequencing results.
How should I check the quality of my sample?
Prior to submitting your purified sample to the SBC for Sanger sequencing, we strongly recommend that you run an agarose gel to confirm the integrity and size of your DNA. On an agarose gel, a tight single band is indicative of good quality DNA. Numerous bands or smearing may indicate:
- degraded DNA
- primer mis-binding (when seen in PCR products)
- primer dimers, resulting from improper purification
- different plasmid conformations (supercoiled, relaxed / linear, nicked)
Additional information to help troubleshoot your agarose gel results can be found by accessing the ThermoFisher website.
In addition, we recommend that you also check the A260/A280 and A260/A230 ratios to assess sample purity. The A260/A280 must be between 1.7 and 1.9.
How should I quantify my DNA prior to submission? Why does this matter?
Incorrect quantitation of your DNA is one of the most common causes of poor sequencing results. We recommend that you submit the appropriate concentration and volume of DNA for your submission type, as described in our submission guidelines. Depending on the sample type or amplicon length, we will adjust the amount of DNA input into the reaction.
Although there are numerous ways to quantify your DNA for Sanger sequencing, we recommend UV absorbance or dsDNA quantification assays such as Qubit or PicoGreen. Nanodrop quantification is quick and dependable for Sanger sequencing purposes, with the major disadvantage being that it overestimates sample concentration due to the presence of contaminants. Please note that the required concentrations listed in our sample guidelines are based on this type of measurement and not fluorometric quantification methods (e.g. Qubit, PicoGreen, QuantIT). Please contact us if the concentrations listed in your sample submission form were not obtained using UV absorbance.
How do I submit my samples?
There are 4 steps required to submit your samples for sequencing:
- Place your order online on dnaLIMS.
- Print out the completed submission form from dnaLIMS.
- Place your samples, primers and completed submission form in a sealed plastic bag.
- Drop off your samples in-person at in the refrigerated dropbox on the main floor of the Pharmaceutical Sciences building (2405 Wesbrook Mall) or contact us for courier delivery information.
Please see our submission guidelines for additional information regarding service type, sample submission requirements and costs.
How do I access dnaLIMS?
You can access our online DNA submission portal here.
You will need to register for an account by clicking on “Create Login Account for dnaLIMS”, and filling in your login and contact information as directed. Once your account is created, you will be able to enter your order and sample information.
I made a mistake entering my sample information. How can I correct it?
You can correct any mistakes online by accessing dnaLIMS.
If you have already mailed or dropped off your samples, please email sanger.submissions@ubc.ca to notify us of your mistake. If your sample has not yet been processed, we will update our information as required.
What can I do to prevent processing delays?
To streamline your submission and ensure your data is provided as quickly as possible:
- Quality-check your samples. More information provided at "How should I check the quality of my sample?"
- Submit samples diluted to the appropriate concentration. More information about quantification can be found at "How should I quantify my DNA prior to submission? "
- If you have less than 48 samples:
- Submit your samples in strip tubes.
- Label the sides of the tubes and not the lids. The label should only contain your initials and sample number.
- Submit samples in the same order specified on your submission form.
- If you have more than 48 samples:
- Submit your samples in a tightly sealed 96-well plate.
- Submit samples by columns, in the same order specified in your submission form.
- Include a maximum of 92 samples per plate, to allow for controls to be included on each sequencing plate.
- If you are using your own primers with our SBC-Prepared service:
- Submit each primer separately from your template DNA.
- Dilute primers to a final concentration of 5 pmol/µL in a 1.5 ml Eppendorf tube.
- Note: several standard sequencing primers are available at the SBC at no extra cost and can be added to sequence your template
How do I access and view my results?
All results can be accessed and downloaded directly from dnaLIMS. Downloaded samples may be analyzed using GeneMapper, FinchTV, Sequence Scanner, ClustalW, and Unipro UGENE.
Our team will notify you by email once your data is available. You may also receive an additional email if we identified any issues in your sequencing results.
How do I know that my samples have been handled appropriately, the sequencing process worked, and that the data I received is from my sample?
How do you keep from mixing up samples or data?
Our team controls and monitors the samples and processes at all stages through the sequencing process.
- During submission:
- Our technologists will check that your submission form contains all required details (sample name, sample concentration, service type, primer name and primer volume) and that your samples are accurately and clearly labelled. They will also visually inspect submitted volumes, comparing these against the printed submission form. Should any clarification be required, someone from our team will contact you before proceeding.
- During the reaction set up and while sequencing:
- We include internal positive and negative controls on all plates to monitor our sequencing process and identify any reagent or instrument issues. If issues are noted in our controls, our team will immediately troubleshoot internal processes to identify the cause. Should troubleshooting cause a delay in data generation, we will notify you as soon as possible.
- During data review and dissemination:
- Our LIMS-based submission system ensures that the client controls sample-related information. Data generated by sequencing the samples is automatically associated with the correct sample, ensuring that client only receives data generated by their samples.
Do you use any controls in your Sanger sequencing pipeline?
The SBC includes positive and negative controls on every plate, which allows us to monitor our Sanger sequencing process, including any issues associated with the 3730XL sequencer.
If issues are noted in our controls, our team will immediately troubleshoot internal processes to identify the cause. Should troubleshooting cause a delay in data generation, you will be notified as soon as possible.
How do I know that my samples are not contaminated while being processed?
We follow numerous separate rigid processes that minimize the risk of cross-contamination. If you suspect that there has been a sample mix-up or that you received results that are not appropriate for the samples submitted, please contact us immediately at sanger.submissions@ubc.ca.
Please refer to our Resequencing Policy for more information.
How do I assess the quality of my sequence?
How do I know if my sequence is of good quality?
Following sequencing, we inspect each sequence prior to data release. Through our analysis software, our technicians evaluate:
- The sequences’ overall sample scores to ensure that the sequence has reliable and usable data.
- The length of the sequence to determine if the sequence generated is longer or shorter than expected.
If any issues are identified in your sequencing results, we will notify you of the issues when we send out your data.
As a general rule, a good quality sequence should have singular peaks that are well defined, sharp and have even spacing between them. The peak height should be high enough that the background noise is minimal in comparison. A sequence failure will often display some of the following characteristics:
- No signal is produced and no peaks are identified.
- Base calls are replaced with N’s.
- All noise in the electropherogram
- The low peak signal in conjunction with baseline noise renders the sample’s signal indecipherable

Figure 1: The sequencing software failed to call any bases with background noise interfering with sample peaks
Why do I need to review the chromatogram file and not just the text file?
Reviewing your chromatogram is an essential part of analyzing and troubleshooting your Sanger sequencing results. As an example, review the text file and chromatogram below. If you only reviewed your data in a text file and blasting this confirms what you were expecting (without ever looking at your chromatogram), you would likely be pleased with your sequencing results. However, your data analysis would be incomplete as a quick view of the overlapping peaks in the chromatogram would suggest your results are not as certain as the sequence initially suggests. As such, your chromatogram acts as a visual control. The Sequencing team will notify you by email if we identified any issues in your sequencing results, which would be apparent in the chromatogram. If your Sanger sequencing result is of lower quality, as shown in the left image of Figure 2, sequencing should be repeated. Please review our Resequencing Policy for more information.
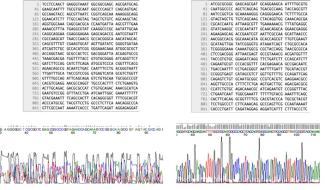
Figure 2: (Left) Text file and corresponding chromatogram trace showing poor quality sequencing data due to spectral pull-up, (Right) Text file and corresponding chromatogram trace showing good quality sequencing data
How do you tell the difference between traces of a PCR product and a plasmid?
Several DNA polymerases such as Taq add a single nucleotide base to the 3‘ end of amplified DNA fragments. The AmpliTaq DNA Polymerase we use during PCR amplification in our Sanger sequencing adds a 3’ adenine overhang to the end of the PCR product. As such, sequenced PCR products are easily identified by their signature adenine peak at the end of the amplicon sequence (Figure 3). This adenine peak is generally larger than its neighbouring peaks and should be well defined, appearing at the end of your sequenced amplicon product (only visible if the amplicon is shorter than the length of sequencing read).
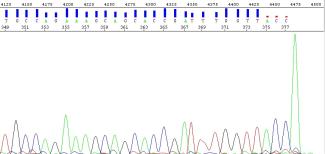
Figure 3: Chromatogram showing the A-tail seen at the end of an amplicon generated with AmpliTaq.
When sequencing a plasmid, no A-tail or abrupt stop is seen at the end of the sequencing product. In a successfully sequenced plasmid, the bases called may seemingly never end (Figure 4). The sequencing software however will stop calling bases once the limits of the 3730XL DNA sequencing instrument are reached. Similarly, longer PCR amplicons, such as those exceeding 1kb, may exhibit comparable characteristics to those seen in a plasmid trace.
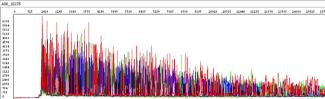
Figure 4: Raw trace file for a successfully sequenced plasmid
My results aren’t what I expected, can you help me understand what went wrong?
What are common reasons for sequencing failures?
Failed or suboptimal sequencing results may be due to a variety of reasons, including:
- The wrong concentration of DNA and/or primer used. The former can be due to inaccurate quantification of the template.
- The template and primer are not compatible (e.g. submission of the wrong combination of template or primer, or incorrect primer design)
- The template is degraded or absent.
- Multiple priming sites or templates are present.
- The presence of contaminating elements interfering with the capillary electrophoresis sequencer. The presence of salt, EDTA, alcohol or detergent in DNA due to insufficient purification of the template can inhibit our 3730XL from producing a sequence. Refer to "What are the effects of contaminants on Sanger sequencing?".
- Secondary structures in your template.
- Technical issues with the 3730XL.
If your samples have failed and you would like help figuring out why we’d be more than happy to assist. Please send an email as soon as you can to sanger.submissions@ubc.ca and we’ll be in touch to troubleshoot.
The SBC will repeat any samples that fail due to our processes or if the 3730XL experiences technical issues but cannot accept responsibility for poor results due to poor quality samples. Please review our Resequencing Policy for more information.
What are the effects of too much or too little DNA in Sanger sequencing?
A ‘ski slope’ effect is often seen in samples that:
- are over- or underloaded with DNA
- use insufficient or too much primer for the sequencing reaction
- use degraded template
As such, it is essential that template material submitted for Sanger sequencing is correctly quantified. We will adjust the amount of DNA input into the reaction, based on the quantifications provided on your sample submission form, as appropriate for the template type and length. Refer to "How should I quantify my DNA prior to submission?" for recommendations on how to quantify your DNA prior to submission for Sanger sequencing.
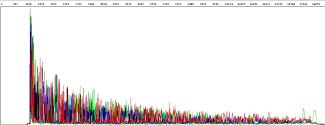
Figure 5: Ski slope effect
What are the effects of contaminants (salt in DNA, EDTA, alcohol, protein, RNA, detergent, cesium) on Sanger sequencing?
Contaminants such as salt, EDTA, alcohol, protein, RNA, detergents or cesium may not affect your results during PCR or restriction digests, however, they can lead to a reduction in resolution when sequencing with capillary electrophoresis. Contaminants can prevent the sequencing polymerase from proceeding further along the template, which may result in numerous short fragments. In extreme cases of contamination, the sequencing signal may be absent altogether.
Samples must be purified prior to submission. Please review the section "How should I prepare my DNA prior to submission for sequencing" for more information. In our experience, failed or suboptimal sequences that occur as a subset within an otherwise successful sequencing plate (e.g. all of the samples from a particular submission), indicate that there may have been an issue in the sample preparation.
Why are there multiple overlapping peaks at the same position? Does this mean my data is unusable?
Overlapping peaks at the same position is often referred to as a ‘mixed trace’ or ‘peaks on peaks’. We will notify you if multiple overlapping peaks are observed in your sequence. Whether or not you can use the data will depend on the goals of your sequencing.
Common explanations for mixed traces are listed below. If unexpected mixed traces occur, we suggest double checking your primers for specificity. In certain cases, the primers may need to be redesigned. Examining your template using agarose gel electrophoresis prior to submission for Sanger sequencing is also recommended, as it will allow you to identify the presence of multiple templates
- Mutations such as insertions or deletions in some of the DNA fragments.
- Multiple primer binding sites on the template.
- Multiple templates or primers present in the reaction.
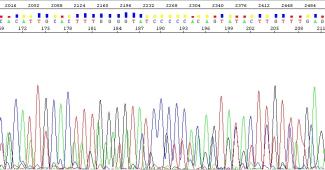
Figure 5: Example of a mixed trace
What are dye blobs, and what are they caused by? Does this mean the data is unusable?
Dye blobs are sharp peaks seen in the chromatogram that are a result of unincorporated dye terminators that were not fully eliminated during the cleanup of the sequencing reaction. At the SBC, we purify after the sequencing reaction to remove unincorporated dye terminators, enzymes and salts that remain following the cycling reaction.
There are a few cases where dye blobs may persist, even following the purification process. For example, samples that failed during the sequencing reaction resulting in weak or no signal may leave a greater proportion of unincorporated dye terminators. This is commonly seen in our non-template controls, where the lack of amplifiable template results in weak signals and a large dye blob. Additionally, the presence of any contaminants such as salt or EDTA in your storage buffers may prevent thorough amplification of the template, resulting in dye blobs.
Dye blobs generally occur in the 70 bp region. In more extreme cases, they may also occur in the 120 bp region. If dye blobs are observed in these regions, we will notify the client about their presence. Even though dye blobs may be observed in your sample, the underlying peaks should allow you to identify your bases manually. However, please contact us at sanger.submissions@ubc.ca if this is not the case.
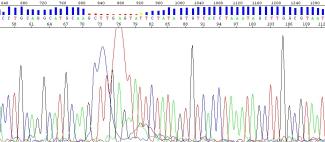
Figure 6: Dye blob occurring in the 70bp region
What are C and G shoulders?
C or G shoulders are described as irregular C or G peaks seen in a chromatogram sequence following sequencing. The end of the C or G peak is not sharp and well defined as seen in a good peak but instead shows an irregularity at the end of the peak where the peak trace extends into a “shoulder”. Please contact us at sanger.submissions@ubc.ca for more information if C or G shoulders are interfering with your data quality.
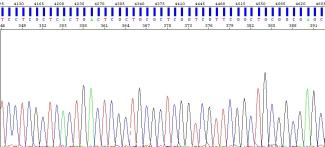
Figure 7: Chromatogram showing good quality sequence, absent of C or G shoulders
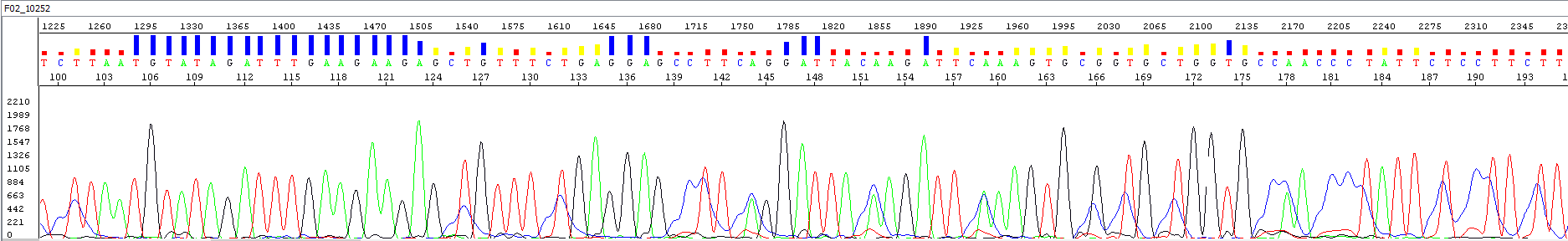
Figure 8: Chromatogram showing the presence of C shoulders
I observed a loss of resolution near the end of my sequence ~800bp. Why does this happen?
As the sequence approaches the resolution limits of the 3730XL sequencer, the probability of errors in base calling increases (Ewing & Green, 1998). Towards the ends of the sequence, the system sometimes has difficulty resolving fragments, which results in lower quality base calling as the base peaks are not well resolved and bleed into each other.
You may notice the following in lower quality regions:
- a reduced quality score
- broader overlapping peaks
- the presence of peaks with no bases called by the sequencing software

Figure 9: Example of high quality, well-defined peaks
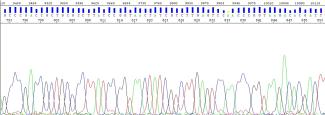
Figure 10: Reduction of quality around the 800th base
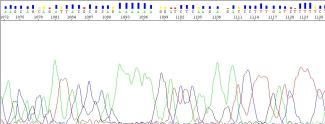
Figure 11: Low-quality peaks near the end of the sequence
Why are the first 15-40 nucleotides in my sequence undeterminable and of poor quality?
The first 15 to 40 nucleotides of a sequence are often poor in quality. The poor quality is due to the Sanger sequencing technology itself, as the technology involves the use of bulky ‘big’ dyes bound to terminator bases. Although these dyes allow the instrument to differentiate between nucleotides during sequencing, the large molecular weights and charges on these dyes cause a shift in migration for the sequencing reaction products undergoing electrophoresis. At shorter fragment lengths, such as those seen in the first 40 nucleotides, these shifts will be more dramatic as the dye composes a greater percentage of the total molecular weight and charge of the sequencing product. Additionally, the presence of unincorporated dyes or incomplete removal of primer dimers following PCR purification may contribute to the messy peaks seen at the beginning of the sequence.
If you require high-quality bases in the first 40 nucleotides of your sequence, we suggest redesigning your primer further upstream of your sequence of interest to avoid any reduction in sequence quality.
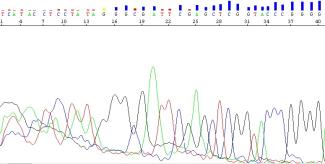
Figure 12: Low-quality sequence in the first 40 bases
My primers produce an amplicon during PCR, but the same primers do not produce a sequence during Sanger sequencing. Why is this?
There are several reasons why the primer you used in PCR amplification failed to produce a DNA sequence:
- There may be residual PCR primers, unincorporated nucleotides and spurious products in your submission.
- Refer to the section "How should I prepare my DNA prior to submission for sequencing" for more information.
- Your submission was inaccurately quantified.
- Refer to the section "How should I quantify my DNA prior to submission?" for more information. We will adjust the amount of DNA input into the reaction depending on the amplicon length.
- Your submitted primer does not meet our primer requirements as stated in our primer recommendations.
- As a sequencing core, we are not able to modify the denaturation, annealing and extension temperatures for each primer.
- PCR is more forgiving than sequencing.
- As PCR is an exponential process using two primers that replicate on opposing strands, amplification can still be seen with suboptimal primers. This is not the case for the amplification stage of Sanger sequencing, which is a linear process using a single primer. Consequently, while primers that do not adhere to our recommendations may work for PCR, these same primers may fail in the sequencing reaction or lead to sub-optimal data.
- Your primers and/or template were submitted in a solution that contains EDTA.
- In our experience, the presence of EDTA often results in downstream sequencing complications.
Resequencing Policy
My results are not what I expected. What should I do? Will I incur a fee for resequencing?
If you feel a mistake was made on our end, we will be glad to resequence the sample to find out if there was an error made, with the following conditions:
- Please contact us as soon as possible at sanger.submissions@ubc.ca to let us know of the potential error.
- We will need to resequence the original sample (template + primer) in its original tube.
- Unfortunately, any sample substitutions or dilutions would not allow us to make conclusions on whether or not a mistake was made on our end.
- If you require us to quantify the failed samples prior to resequencing, this option is available at an additional cost. Otherwise, we can resequence based on your submitted quantification.
- If the resequenced reaction is successful, then a mistake was clearly made on our end and you will not be charged for the resequencing.
- If your sample fails a second time, then we must assume the problem was with your sample and you will be charged for the original sequence as well as for the resequencing due to the extra effort involved.
Please note: two "identical" sequencing reactions may differ slightly but noticeably. Resequencing may produce marginally better or worse results when compared to the first run.
References
Choi, J. S., Kim, J. S., Joe, C. O., Kim, S., Ha, K. S., & Park, Y. M. 1999. Improved cycle sequencing of GC-rich DNA template. 1999. Experimental Molecular Medicine, 31, 31(1) 20-24. DOI: 10.1038/emm.1999.3
Ewing, B., & Green, P. 1998. Base-Calling of Automated Sequencer Traces Using Phred II Error Probabilities. Genome Research, 8, 186-194. Cold Spring Harbor Laboratory Press. ISSN 1054-9803-98
Shapter, F. M., & Waters, D. L. 2014. Genome Walking. Methods Molecular Biology, 1099, 133-146. DOI: 10.1007/978-1-62703-715-0_12.