Submission Overview
Step 1: Prepare samples specifically for the preferred service level
Step 2: Enter your order request only using dnaLIMS, and submit samples with a printed copy of your request form together in a sealed bag.
Step 3: Drop off in-person in the refrigerated dropbox on the main floor of the Pharmaceutical Sciences building (contact us for details), by campus mail, or by courier to:
Sequencing and Bioinformatics Consortium
University of British Columbia
Room 3124 - PharmSci Building
2405 Wesbrook Mall
Vancouver, BC V6T 1Z3
Submission Deadlines:
- 9:00 am for SBC_Core-Prepared samples
- 2:00 pm for all other sample types
Submission Guidelines
Following these guidelines will ensure that your samples are processed without delay.
Preparing Your Order
A maximum of 92 samples can be submitted in each order to allow for the inclusion of controls. Your order request must be submitted online using dnaLIMS prior to physically submitting your samples:
- Orders ≤ 48 samples: submit in 8-strip PCR tubes
- Label the side of the tubes with the sample number
- Orders > 48 samples: submit in 96-well PCR plate
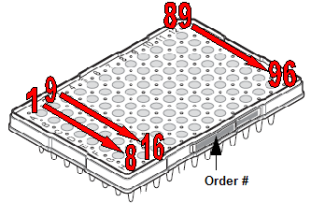
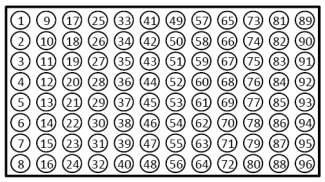
- Submissions in 96-well plates must be in the following orientation:
Note: We can provide you with strips or plates if needed, please contact us at sanger.submissions@ubc.ca for details.
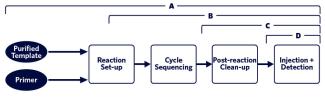
The SBC offers four service levels for Sanger sequencing, each with its own submission guidelines:
Template and Primer Requirements
**Important: Templates and primers must be resuspended in molecular biology grade water or 10 mM Tris pH 7-8 with no EDTA, as EDTA will interfere with the ion concentration in the sequencing reaction.